Ingesting MERFISH data with Panpipes
This tutorial guides you through the ingestion of MERFISH datasets using Panpipes. The workflow describes the steps run by the pipeline in greater detail.
For all the tutorials, we will append the --local command which ensures that the pipeline runs on the computing node you’re currently on, namely your local machine or an interactive session on a computing node on a cluster.
Create directories and download data
Create a main spatial directory and inside it, ingestion and data.
mkdir spatial
cd spatial
mkdir ingestion ingestion/data
In this tutorial, we will use a mouse brain dataset that you can download here (Slice 1, Replicate 1).
After downloading the data, your spatial folder should look as follows:
spatial
├── ingestion
│ ├── data
│ ├── datasets_mouse_brain_map_BrainReceptorShowcase_Slice1_Replicate1_cell_by_gene_S1R1.csv
│ ├── datasets_mouse_brain_map_BrainReceptorShowcase_Slice1_Replicate1_cell_metadata_S1R1.csv
│ ├── images
│ └── datasets_mouse_brain_map_BrainReceptorShowcase_Slice1_Replicate1_images_micron_to_mosaic_pixel_transform.csv
Please note, that the data folder structure needs to be structured as expected by the squidpy.read.vizgen function.
Edit submission and yaml file
Create a submission file like the one we provide and save it inspatial/ingestion. For this tutorial, you can use the provided submission file. Detailed information about the submission file is provided in the usage guidelines
Next, in spatial/ingestion call panpipes qc_spatial config (you potentially need to activate the conda environment with conda activate pipeline_env first!). This will generate a pipeline.log and a pipeline.yml file.
Customize the pipeline.yml or simply replace it with the one we provide. Make sure to specify the correct path to the submission file. If you’re using the provided example yaml file, you potentially need to add the path of the conda environment in the yaml.
Run Panpipes
In spatial/ingestion, run panpipes qc_spatial make full --local to ingest your MERFISH datasets.
After successfully running the pipeline, the ingestion folder looks as follows:
ingestion
├── data
├── figures
│ └── spatial
│ ├── histograms.mouse_brain.png
│ ├── spatial_spatial_n_genes_by_counts.mouse_brain.png
│ ├── spatial_spatial_total_counts.mouse_brain.png
│ ├── violin_obs_n_genes_by_counts_sample_id.mouse_brain.png
│ ├── violin_obs_total_counts_sample_id.mouse_brain.png
│ └── violin_var_total_counts.mouse_brain.png
├── logs
│ ├── make_mudatas_mouse_brain.log
│ ├── qcplot.mouse_brain.log
│ └── spatialQC_mouse_brain.log
├── qc.data # MuData with QC metrics
│ └── mouse_brain_unfilt.h5mu
├── tmp # MuData without QC metrics
│ └── mouse_brain_raw.h5mu
├── pipeline.log
├── pipeline.yml
├── sample_file_qc_spatial.txt
└── mouse_brain_cell_metadata.tsv # Metadata, i.e. .obs
In the qc.data folder, the final MuData object with computed QC metrics is stored. The MuData object without QC metrics is also available and stored in the tmp folder. The metadata of the final Mudata object is additionally extracted and saved as tsv file, mouse_brain_cell_metadata.tsv.
Using the provided example yaml file, the first rows and columns of the mouse_brain_cell_metadata tsv file look as follows:
spatial:fov |
spatial:volume |
spatial:min_x |
spatial:max_x |
spatial:min_y |
spatial:max_y |
|
|---|---|---|---|---|---|---|
110883424764611924400221639916314253469 |
0 |
432.1414246586824 |
151.53048381581905 |
161.5960840769112 |
4264.619631592184 |
4278.0332319401205 |
With the plots in spatial/ingestion/figures/spatial you can now decide on cutoffs for filtering. The plots include visualizations of the spatial embeddings, as well as violin plots, and histograms:
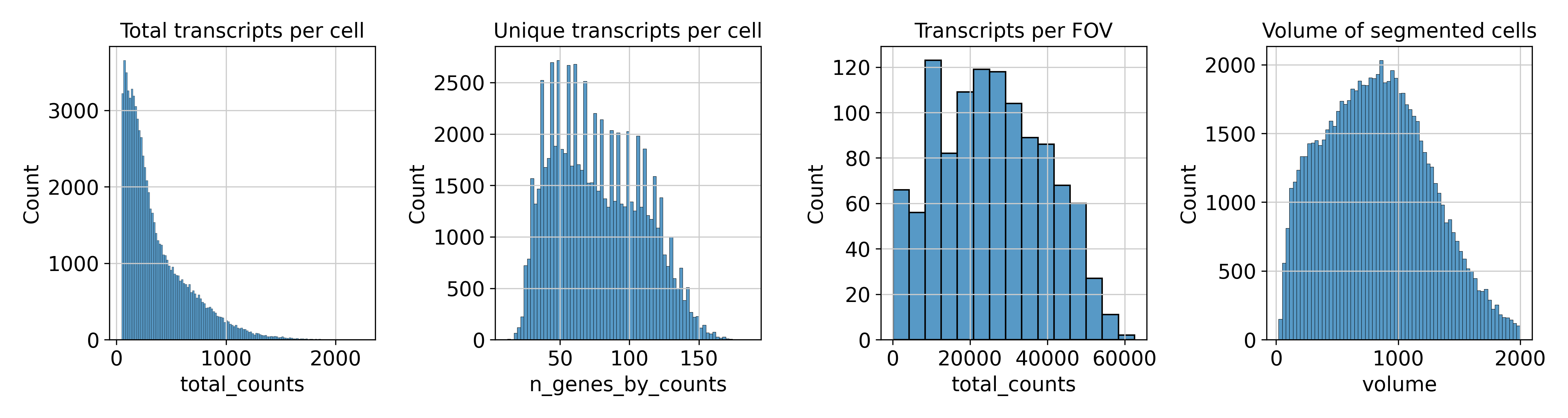
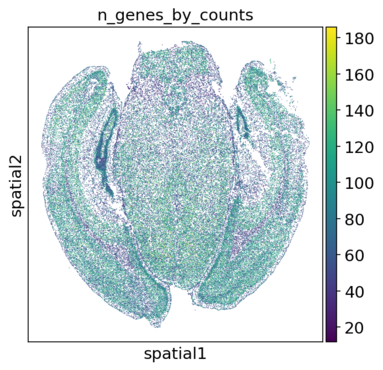

Next: filtering and preprocessing using panpipes preprocess_spatial
Note: In this workflow, we have decided to process individual ST sections instead of concatenating them at the beginning, as you saw for cell-suspension datasets. This is because the workflows for processing multiple spatial transcriptomics slides (especially concerning normalization, dimensionality reduction, and batch correction) are still experimental. With panpipes, you can group multiple samples and process them one by one with the same choice of parameters. In the future we will implement the advanced functionalities of SpatialData to deal with multi-sample ST datasets.
Note: We find that keeping the suggested directory structure (one main directory by project with all the individual steps in separate folders) is useful for project management. You can of course customize your directories as you prefer, and change the paths accordingly in the pipeline.yml config files!